CoxBase Help Page
Coxbase is an online platform for epidemiological surveillance, visualization, analysis and typing of Coxiella burnetii genomic sequence
This help page is subdivided into functional sections based on the navigation features
Distribution map
The Distribution categorizes isolates per country. The number signifies the total number of Isolate recovered in the country.
Clicking the ball icon will reveal a card pop up with further details as shown in the image below
The numbers are color coded based on ranges shown on the top right corner

Two further links are available on the pop up card: Isolates table and Dashboard (Visualizaton of the country dataset according to different meta data categories)
Query
The query feature is available for isolate discovery and comparison. 3 types of queries are available currently: MLVA, MST and Primer sequence for typing methods
For the MLVA queries, there are 14 input areas for the 14 MLVA loci used by Frangoulidis et al 2015 paper. At least one input field must be entered for a succesful query.
When a perfect match is found only one profile is returned else several profiles are returned.
The view profile entries on the last column can be used to further explore the isolates with similar profiles
For the MST queries, there are 10 input fields for each of the 10 interspacer regions described by Glazunova et al 2005.
The MST query are meant to discover the group ID of an isolate based on unique combination of the the different interspacer region
The first column in the result table is the MST ID column
Analysis
There are 5 possible typing analysis on CoxBase presently. The MLVA typing analysis, MST typing analysis, IS1111 typing, AdaA gene and Plasmid. All typing methods require either a sequence upload (recommended) or sequence input in the text area.
NOTE: Sequence must be in Fasta format
After the sequence has been uploaded. Run the typing analysis by clicking on the submit button.
Analysis Results
MLVA
A result table with details of the in silico analysis such as the product length, flank length, the used repeat size is presented with the calculated repeat number highlighted as the base row as shown below
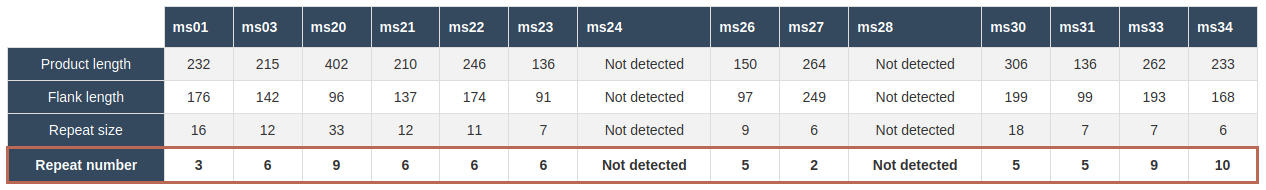
NOTE: Not detected signifies the amplicon locus was not detected in silico, there could be several reasons for this but the most likely is an incomplete assembly
3 links are provided above the result table, as shown below
- Result table - permanent link that point to the result of this analysis and can be saved or bookmarked for further consultation
- Submission details - points to the page that contain details of the particular submission and it includes a 36 unique alphanumeric which is the Submission ID. This can be used to retrieve the analysis historically
- Phylogenetic analysis - takes the user to a page with a link to query the MLVA database with the calculated repeat numbers
MST
The result table shows the allele type for each of the 10 inter spacer region. The MST group can be discoverd by clicking on phylogenetic analysis and submiting the result to the MST database as shown below

The column headers are the spacer type and the values below are the detected alleles.
NOTE: New Cox56 signifies the detected allele is not in the MST alleles database. We recommend a close examination of this allele sequenece by clicking on the button so as to distinguish between a true SNP variant and a false variant due to missing or unannotated nucleotides. A BLAST result annotation constructed with Blaster JS will be pulished under the result table for futher clarification.
IS1111
The IS1111 method is based on the localization of IS1111 regions. The result table contains alternating columns of IS element and the state ( status if it's present or absent )
 indicates present
indicates present
 indicates absent
indicates absent
Retrieval
The retrieval feature is available for users who would like revisit an historically concluded analysis.
How it works:
Every submitted job has a unique submission ID which can be found on the submission details page as shown below:

